Wayward Ways: New Study Reveals How the Nucleus Travels
Quick Summary
- Proper positioning of the nucleus in the cell is vital for many biological processes
- Nuclear migration is controlled by a bridge of proteins known as the LINC complex, built by SUN and KASH proteins
- By mutating genes associated with these proteins, the team created a blueprint of interaction for nuclear migration
From the start of life, the proper positioning of the nucleus in the cell is essential in many developmental processes, including fertilization, cell migration and neuronal and muscle development. However, many questions remain regarding how exactly the nucleus moves and repositions itself in the eukaryotic cell.
A new study from UC Davis published in Current Biology helps define this process for researchers. Professor Daniel Starr, Department of Molecular and Cellular Biology, and his colleagues at UC Berkeley and the University of Minnesota have revealed how two groups of proteins responsible for nuclear positioning—called SUN and KASH, respectively—function, providing researchers with a blueprint of interaction.
“It’s a nice three-way collaboration between worm genetics, mammalian tissue culture experiments and computer modeling,” said Starr.
The nuclear bridge
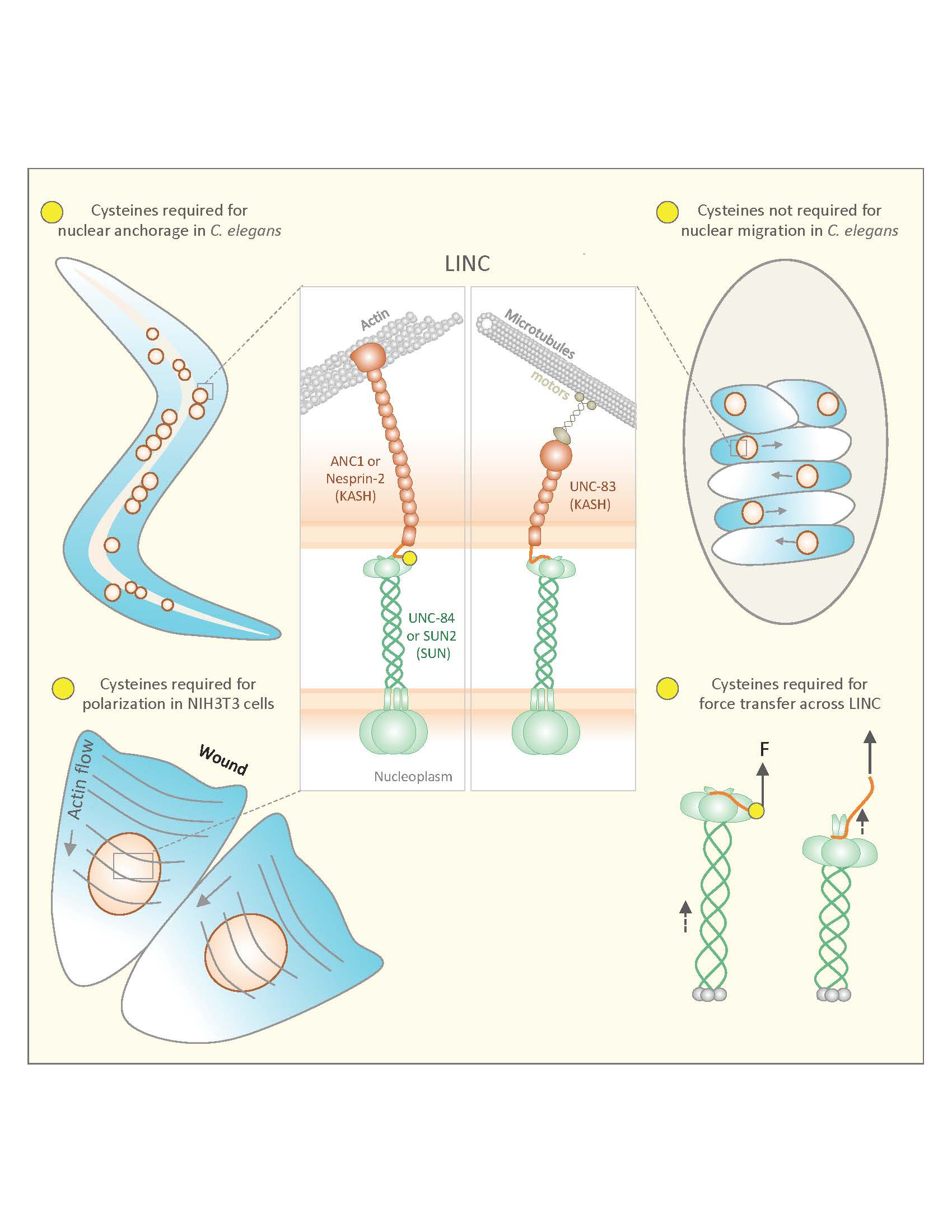
Inside the cell, the nucleus’ anchorage and movement is controlled by a biological bridge of proteins called the linker of nucleoskeleton and cytoskeleton (LINC) complex. This bridge of SUN and KASH proteins connects structures inside the nucleus to those outside of the nucleus. According to Starr, the LINC complex is conserved across all eukaryotic species, indicating that it’s a fundamental part of eukaryotic biology.
In mammals, SUN and KASH protein groups are integral to many biological functions, including skin, retina, hair cell, sperm and muscle development and homologous chromosome pairing. Their dysfunctions can lead to sterility, blindness, autism and cancer, to name a few negative impacts.
Previously, structural biologists identified the LINC complex’s structure, predicting the functions of the SUN and KASH proteins comprising it. According to Starr, the SUN proteins in this complex are positioned along the inner nuclear membrane while KASH proteins sit on the outer nuclear membrane. These two protein groups interact with each other to form the LINC complex and to achieve anchorage or movement.
“The LINC complex evolved at the same time as the nuclear envelope and was probably present in the last eukaryotic common ancestor,” said Starr. “Even today, we don’t understand how this machine is put together or how it’s regulated during development.”
Worms, mice and computers
Using CRISPR/Cas9 gene editing tools, Starr created mutant versions of the worm C. elegans, focusing on parts of the genome responsible for expression of SUN and KASH proteins. He then observed how these proteins interacted during the worms’ development.
Starr and colleagues found that the worms used a KASH protein called UNC-83 for movement and another called ANC-1 for anchorage in the cytoskeleton. Both UNC-83 and ANC-1 interacted with a SUN protein called UNC-84 to achieve movement and anchorage, respectively.
“So earlier in development, UNC-84 needs to interact with UNC-83 to move the nucleus along the cell’s microtubules, and then later in development, UNC-84 needs to interact with ANC-1 to anchor the nucleus,” said Starr.
Additionally, the team confirmed the importance of cysteine residues, which form bonds between SUN and KASH proteins, in the LINC complex. The Starr lab hypothesized that if these residues existed in the worm, regulation of the SUN and KASH bond could be the key to controlling developmental switches between nuclear migration and anchorage. Starr’s lab found that mutating the conserved residues disrupted SUN and KASH function in worms.
According to Starr, University of Minnesota colleagues tested the same hypothesis but in mammalian tissue cell cultures. When they mimicked mutations made in Starr’s lab, they discovered mammalian cells also exhibited disruptions in nuclear positioning. UC Berkeley colleagues then used computer simulations to predict the interactions of all the atoms involved in this cellular process. The models confirmed the importance of cysteine’s role in mediating mechanical forces required to move and anchor nuclei.
What’s calling the shots?
While the study creates a blueprint of the nuclear migration and anchorage processes, researchers still don’t know how the cell regulates these decisions.
“There’s a bunch of potential players in the nuclear envelope that could be doing that,” said Starr, noting several enzymes and chaperone proteins in the cell. “The structure predicted that the cysteines were important and we tested and showed that it is important. Now the question is, how is the cell regulating the interactions? And what is the cell doing to make and break those bonds?”
The research was supported by the National Institutes of Health, National Science Foundation, Natural Sciences and Engineering Research Council of Canada, Dystonia Medical Research Foundation and American Cancer Society Illinois Division.
